
Introduction
Neoantigen prediction based on 3D
genome information and Deep Sparse
Learning.
The mutations of cancers can encode the seeds of their
own destruction, in the form of T cell recognizable immunogenic
peptides, also known as neoantigens. In this work, we for the first
time considered the original DNA loci of the neoantigens in the
perspective of 3D genome which contributes to better
immunogenicity prediction. We employed the 3D genome information along
with an ensemble pMHC-I coding strategy, and developed a group feature
selection based deep sparse neural network model (DNN-GFS) that is
optimized for neoantigen prioritization. This webserver implements the
DNN-GFS as well as other machine learning methods. We hope this work
provides a new perspective towards more accurate neoantigen prediction
which eventually contribute to personalized cancer immunotherapy.
Workflow of neoantigen therapy supported by 3D genome information.
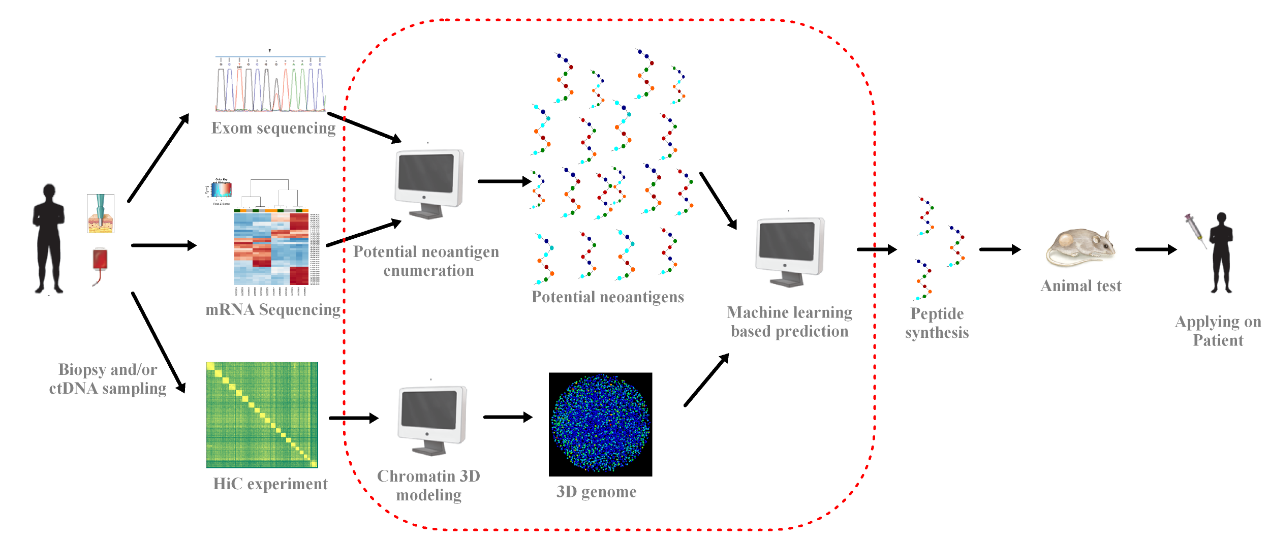
Left to right: tumor sample collection from patient; Whole-exome sequencing and mRNA sequencing for somatic mutations calling and gene expression estimation (whether the mutated DNA is expressed into mRNA and could potentially be translated into protein/peptide) respectively; Hi-C data curation to obtain 3D genome information; candidate peptides determined by NGS are generated and by combining 3D genome information immune-positive peptides are predicted machine learning methods; the top ranked peptides are screened by conducting animal experiments; the final peptide penal can be applied back to the target patient. This work aims to solve the tasks within the dashed red frame.
Citation:
Yi Shi+* , Zehua Guo+, Xianbin Su+, Luming Meng*, Mingxuan Zhang, Jing Sun, Chao Wu, Minhua Zheng, Xueyin Shang, Xin Zou, Wangqiu Cheng, Yaoliang Yu, Yujia Cai, Chaoyi Zhang, Weidong Cai, Lin-Tai Da*, Guang He*, Ze-Guang Han*, DeepAntigen: A Novel Method for Neoantigen Prioritization via 3D Genome and Deep Sparse Learning, Bioinformatics, DOI: 10.1093/bioinformatics/btaa596, Published on June 27th, 2020.
Yi Shi+, Xianbin Su+, Kunyan
He, Binghao Wu, Boyu Zhang, and Ze-Guang Han*. Chromatin accessibility contributes
to simultaneous mutations of cancer genes. Scientific Reports.
6:35270. 2016.
The neoantigen prioritization work was inspired
by this previous discovery that somatic co-mutations from the same
sample (or even the same single cell) tend to locate proximately in 3D
genome, forming somatic co-mutation hotspots (SCH) in chromatin 3D
space.
Contact:
Yi Shi, yishi[at]sjtu.edu.cn
Zehua Guo, guozehua[at]sjtu.edu.cn